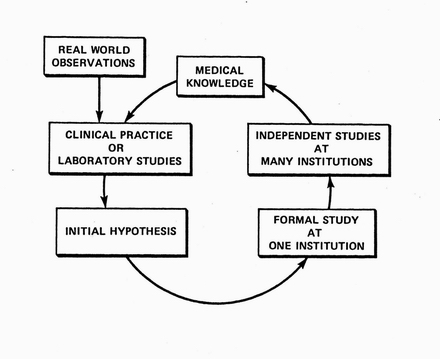
The cycle of discovery in the real world. Informed by existing medical knowledge, clinical researchers or lab-based researchers formulate hypotheses that are preliminarily tested in small studies. Promising hypotheses are then indepently studied at more institutions. The resulting knowledge further guides the formulation of new hypotheses.
Imagine, as I did in 1976 when I came to Stanford, a future world in which every scrap of patient data was collected on computers: every symptom, every lab test or x-ray, every treatment. (That, of course, is the world we are headed for today.)
In those days patient info was scribbled on scraps of paper bound into a manila folder.
Next, imagine a computer tirelessly looking through that data trying to discover new relationships. What might it find?
What are the causes of disease? What are the symptoms? What are the best treatments? Are there habits associated with good health and long life? The computer would keep track of what it found and try to verify it on other massive patient databases.
It might announce its discoveries in the morning when the researchers gathered for their coffee. (Or, it might just undertake unauthorized research projects as in my AI Awakens .)
That was the image behind my Stanford PhD thesis research, called the RX Project.
I designed and programmed the project from 1976 to 1981 and then, as a (research associate) Principal Investigator, I led a team of students, statisticians, and computer scientists to refine and test it until 1986, when I returned to clinical practice. (Now, I'm back on campus again, trying to integrate AI and cognitive neuroscience — if such an integration is possible.)
By the way, my co-PI and head of my PhD thesis committee was Professor Gio Wiederhold, a database expert, who at age 80 continued teaching in the Stanford CSD. (Gio died on December 26, 2022 at age 86. He mentored 36 PhD students and published over 400 papers.) Also on my thesis committee were Professors Edward Feigenbaum (AI), Bruce Buchanan (AI), Stanley N. Cohen (molec bio), Byron Wm Brown (biostatistics), and James Fries (immunology/informatics).
RX was an early example of data mining or exploratory data analysis under AI control. (The pioneers are the guys with the arrows in their backs! At that time nobody was doing machine learning from big data; now everybody is!) RX included a built-in knowledge base of clinical information. It also included a “robot statistician” that it used to design its studies once a promising hypothesis had been discovered. Its discoveries would automatically be incorporated as new machine-readable knowledge.
The database that RX mined was the 1700 patient, multi-decade clinical database ARAMIS , managed at Stanford by Professor Fries for the American Rheumatism Association.
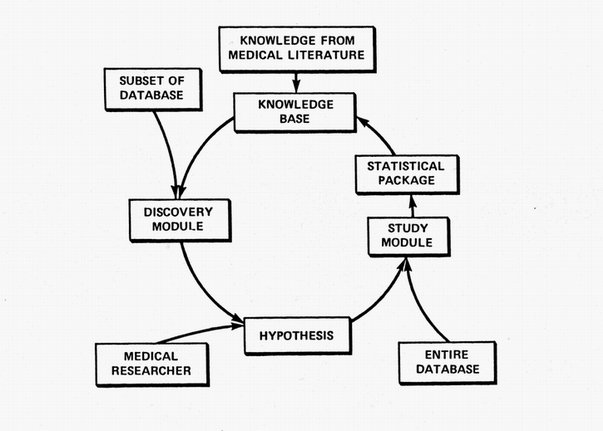
The ultimate goal of the RX Project was to continually and automatically crank out medical knowledge. Its never-ending cycle was intended to emulate the cycle of clinical discovery shown in the first figure. Its Discovery Module suggested hypotheses based on data subsets. Those candidates were then studied comprehensively on the entire database using a robot statistician. The resulting new knowledge was then added to its existing archive of knowledge where it was used to help guide and constrain new studies. The medical researcher might become an expensive redundancy.
RX rediscovered several interesting and important causal relationships — principally drug side-effects— and the system was widely studied by medical computer scientists in the United States, Europe, and Japan.
In 1985 I was among three scientists awarded the Toyobo Foundation's (Japan) top prize for bioinformatics.
Below are sections from the publications that most completely described this work.
(These publications are not online except here. This was a decade prior to the web era.)
(Machine learning from big data is now a hot topic, but this field remained unexplored for decades. You'll find out why in a minute.)
Automated Discovery: the RX Project — Publications
First, an abstract of my first big paper in 1982 based on my Stanford PhD research that appeared in Computers in Biomedical Research:
Next: the big paper — it's 24 pages long, so I've divided it into two pieces (scanned by hand.) This initial piece gives an overview.
Automated Discovery: the RX Project (first 9 pages, 8mb)
Next: the remaining 15 pages (15 mb) for the truly motivated.
Automated Discovery; the RX Project: the last half
Why wasn't this work replicated elsewhere? Stanford had a unique combination of resources that existed nowhere else in that era —
(Even with the Decsystem-20's leading edge hardware, a discovery run would take 3 cpu days in batch mode!)
Historical Note
The ARAMIS clinical database contained "vast megabytes" of data that could most easily be transferred from its source (a campus IBM 370) to the SUMEX-AIM Arpanet Decsystem20 on magnetic tape (hand carried by me.)
But one day, in the early 1980s all that changed.In this heady atmosphere, startups were everywhere. One of the best known was Cisco Systems, which commercialized the routers and switches that made computer networking and data transfer possible. Cisco was founded by my CSD fellow grad student Len Bosack with his MBA wife, Sandy Lerner. So, suddenly data tranfer in the early 80s became easy.
While Len and Sandy made the big bucks at Cisco, it's been acknowledged that priority of invention for the multiprotocol router must go to Bill Yeager, then a researcher at SUMEX-AIM and my frequent running buddy. So, Bill should get the glory, although Len got the money. (I would still occasionally chat with Len when we were buying components at Fry's Palo Alto (R.I.P. 2020.)
RX's most important medical discovery was that prednisone elevates cholesterol.
This was published in the Annals of Internal Medicine in 1986.
Prednisone elevates Cholesterol: Abstract
The entire 1986 Annals paper (14 mb)
The Annals paper is a detailed description of the statistical methods used by RX to quantitate this association in the face of missing and sporadic data.
One of our key insights was to do all detailed analyses first on individual patients over time (before combining those results across patients (weighting by the inverse of the variances.) This is a theme I hear repeatedly nowadays from the neuroscientists. People are so different that high signal to noise ratio can best be achieved by looking at each individual.) In the world of clinical medicine that means using large clinical databases (EMRs) gathered over years with many data points on each patient.)
One of many AI pieces of the RX Project was its representation and use of causal relationships.
Causal relationships (CR's) are a crucial part of our knowledge of the world. Here are some examples. Drinking too much alcohol can cause headaches. Exercise promotes longevity. Burning fossil fuels increases atmospheric CO2.
The main task of RX was to learn CRs (ie infer, store, and use them) from the clinical data. Once RX derived a putative causal relationship from its database, the next step was "learning it" by incorporating the new CR into its knowledge base of clinical medicine.
Its methods are explained here Representation of Empirically Derived Causal Relationships,
and here Modeling and Encoding Clinical Causal Relationships in a Knowledge Base
Before starting work on machine discovery of causal relationships, I first worked on the well known MYCIN Project, which automated infectious disease therapy.
In this task, MYCIN was shown by us to be comparable in accuracy to infectious disease specialists.
Here are more reports on the RX Project and work inspired by it, curated by Professor Gio Wiederhold. (Note: while the list is accurate, many links are unmaintained.)
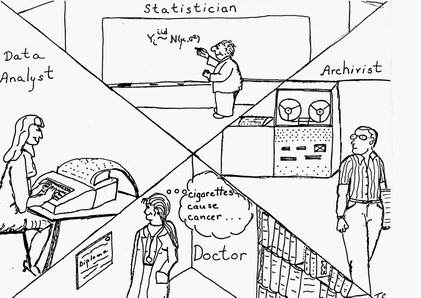
The AI effort in RX also included automating the tasks of the statistician (selecting statistical methods and interpreting data sets) and the physician/epidemiologist (choosing hypotheses, controlling confounders, and "publishing" (incorporating) results.
Note, in the figure above, the doctor is female — somewhat more unusual in 1979. (The artist, my ex-wife, was not about to draw all the highly-paid professionals as being male!)
Also, note the quaint computing equipment in Triona's drawing — from the mag tape reader to the key punch machine. All my programs from my first job (for George Danzig in OR at Cal in 1965 on the campus IBM 360 up to and including my LISP class with John McCarthy in 1976) used punch cards!) The lone exception was my first programming experiences in 1962 at Cal on an IBM 1620 actually seated at the console and entering assembly language programs.
RX's notion of causality was one promulgated by (recently deceased, Stanford superstar) Professor Pat Suppes.
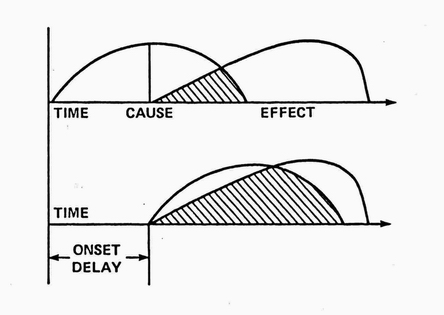
In RX's parlance, a causal relationship was defined as a nonspurious, time-lagged correlation between two variables. Nonspuriousness means that the correlation is not, in fact, caused by some third (confounding) variable(s.) In practice, this can be difficult to establish in biological systems even with lab experiments and randomized, clinical trials . RX's Discovery Module used time-lagged, nonparametric correlations to suggest initial hypotheses.
A key task of RX's Study Module was to use the RX Knowledge Base to attempt to control for known confounding variables, however causally remote, using directed acyclic graphs and multiple regression.